Learning the apply family of functions
One of the greatest joys of vectorized operations is being able to use the
entire family of apply functions that are available in base R.
These include:
apply
by
lapply
tapply
sapplyapply
apply applies a function to each row or column of a matrix. It's a convenient
way to get marginal values. It follows this syntax: apply(object, dimension,
function).
m <- matrix(c(1:10, 11:20), nrow = 10, ncol = 2)
m [,1] [,2]
[1,] 1 11
[2,] 2 12
[3,] 3 13
[4,] 4 14
[5,] 5 15
[6,] 6 16
[7,] 7 17
[8,] 8 18
[9,] 9 19
[10,] 10 20
# 1 is the row index
# 2 is the column index
apply(m, 1, sum) # row totals [1] 12 14 16 18 20 22 24 26 28 30
apply(m, 2, sum) # column totals[1] 55 155
apply(m, 1, mean) [1] 6 7 8 9 10 11 12 13 14 15
apply(m, 2, mean)[1] 5.5 15.5
There are convenience functions based on apply: rowSums(x), colSums(x),
rowMeans(x), colMeans(x), addmargins.
by
by applies a function to subsets of a data frame.
by(prod.long$milk, prod.long[, "test"], summary)prod.long[, "test"]: 1
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
10.40 26.00 33.00 33.41 40.90 59.20 135
--------------------------------------------------------
prod.long[, "test"]: 10
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
5.20 16.50 21.60 21.78 25.90 45.30 332
--------------------------------------------------------
prod.long[, "test"]: 2
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
9.40 27.70 34.40 35.05 41.90 60.30 141
--------------------------------------------------------
prod.long[, "test"]: 3
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
3.80 25.20 32.00 32.86 39.00 64.50 158
--------------------------------------------------------
prod.long[, "test"]: 4
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
10.00 24.00 30.50 31.29 38.58 57.60 174
--------------------------------------------------------
prod.long[, "test"]: 5
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
10.10 22.10 29.80 29.74 36.10 54.20 199
--------------------------------------------------------
prod.long[, "test"]: 6
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
10.00 22.30 28.40 28.43 34.60 54.80 219
--------------------------------------------------------
prod.long[, "test"]: 7
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
9.70 20.22 26.55 26.71 32.70 52.50 234
--------------------------------------------------------
prod.long[, "test"]: 8
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
4.20 19.08 25.20 24.95 30.13 46.90 252
--------------------------------------------------------
prod.long[, "test"]: 9
Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
5.00 18.20 23.00 23.10 27.05 45.50 285
Exercise 1
Using by, what is the mean milk production for each monthly test? (use the
prod.long dataset)
Solution
by(prod.long$milk, prod.long[, "test"], function(x) mean(x, na.rm = TRUE))tapply
tapply applies a function to subsets of a vector.
tapply(health.wide$age, health.wide$lactation, mean) 1 2 3 4 5 6 7
2.234899 3.372519 4.447423 5.574000 6.663043 7.457143 8.685714
8 9
9.825000 10.100000
tapply() returns an array; by()returns a list.
lapply (and llply)
What it does: Returns a list of same length as the input. Each element of the output is a result of applying a function to the corresponding element.
my_list <- list(a = 1:10, b = 2:20)
my_list$a
[1] 1 2 3 4 5 6 7 8 9 10
$b
[1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
lapply(my_list, mean)$a
[1] 5.5
$b
[1] 11
sapply
sapply is a more user friendly version of lapply and will return a list of
matrix where appropriate.
Let's work with the same list we just created.
my_list$a
[1] 1 2 3 4 5 6 7 8 9 10
$b
[1] 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
x <- sapply(my_list, mean)
x a b
5.5 11.0
class(x)[1] "numeric"
replicate
An extremely useful function to generate datasets for simulation purposes.
replicate(10, rnorm(10)) [,1] [,2] [,3] [,4] [,5]
[1,] -0.18296801 -0.07941687 0.556866478 0.32202953 0.49421349
[2,] 1.02546237 0.23094961 -0.027725896 -1.33510987 -0.36789050
[3,] 0.61763429 0.27703658 -0.633289121 0.97164947 -1.24375323
[4,] -0.78905915 0.32296508 0.590272159 1.31506326 1.41135827
[5,] -0.51034496 0.63508227 2.055402761 -0.89659294 -0.28714015
[6,] -0.37624329 0.78419520 -1.205187780 -0.29507644 1.32026833
[7,] 0.08537509 1.86086714 -0.970240977 0.07661003 -2.25080168
[8,] -0.14789145 1.57162130 -0.007669854 -0.05094117 -0.02900168
[9,] -0.26309675 0.86304561 -0.733189759 -0.03084748 -1.61350051
[10,] -0.05618467 0.89226587 -1.266500283 -0.59140376 1.02791954
[,6] [,7] [,8] [,9] [,10]
[1,] 0.4426889 0.03526615 -0.90270328 -0.26814034 0.5401839
[2,] 1.0911277 -0.08477494 0.39941108 0.82335054 0.2204077
[3,] 0.5253514 0.19089756 0.97723529 -1.72851775 0.1809154
[4,] -0.9069135 -0.79663168 -0.69463126 0.05781333 0.6269190
[5,] 1.0669793 -3.08790009 0.69193494 -0.67787888 -1.0643246
[6,] -1.3896023 -0.30786629 -0.82441093 -1.09917333 0.6365318
[7,] 1.1329454 -1.44655598 0.35700710 0.58868047 -1.1802607
[8,] -1.1190632 -0.90202972 0.59884816 -0.57900125 0.4512936
[9,] 1.1162238 1.03717790 0.41516267 -0.14242241 0.1062566
[10,] 1.9634481 -1.18035318 -0.02035708 -0.81575567 -0.5582405
replicate(10, rnorm(10), simplify = TRUE) [,1] [,2] [,3] [,4] [,5]
[1,] -0.54746727 0.71188242 -0.7692378 0.20776940 -1.1911955
[2,] 0.17129389 1.17065806 0.7341176 -0.19784111 1.4999466
[3,] 1.09769758 -0.04547800 0.5835762 -1.98329516 -0.5509973
[4,] 0.49039424 -1.58614965 1.7075941 -0.48315885 0.7405964
[5,] 0.33509901 0.38512102 -0.7831752 0.28508535 0.2430618
[6,] 0.92131753 -0.15772011 -0.4985644 0.00217137 0.5447922
[7,] -0.01576046 0.11304573 -0.6274871 -0.25288329 -0.2649672
[8,] 1.73799532 0.02527156 0.3225937 -1.67327665 -1.0400179
[9,] -0.97862588 0.62199262 1.0316596 -0.62823981 0.3306254
[10,] -0.66175684 -0.53292217 -1.0175170 -1.70367106 0.8996907
[,6] [,7] [,8] [,9] [,10]
[1,] 0.20172680 0.39767985 -1.4565158 0.343714295 0.608689123
[2,] -0.09933205 -0.82269883 0.2607614 -0.913100277 -0.731100767
[3,] -1.64026535 -1.34769622 0.1357663 -1.348551922 -1.287339421
[4,] 0.46878069 -1.44551108 1.8891981 -0.456134156 0.001269542
[5,] -0.13024399 0.79899685 -1.0435204 -0.413426122 -1.458832823
[6,] -1.02578585 0.01650778 0.3144996 -1.103240123 -0.650437291
[7,] -0.98625566 -1.01259025 2.4682385 -0.585127293 -1.051062421
[8,] -0.22434056 0.31500438 -0.1349445 -0.082358386 1.120358846
[9,] 1.60622386 -1.34955072 -0.1424986 -0.454433715 -1.773935613
[10,] 0.44464687 -0.38118115 1.0751461 -0.009772592 1.328939639
The final arguments turns the result into a vector or matrix if possible.
mapply
Its more or less a multivariate version of sapply. It applies a function to
all corresponding elements of each argument.
example:
list_1 <- list(a = c(1:10), b = c(11:20))
list_1$a
[1] 1 2 3 4 5 6 7 8 9 10
$b
[1] 11 12 13 14 15 16 17 18 19 20
list_2 <- list(c = c(21:30), d = c(31:40))
list_2$c
[1] 21 22 23 24 25 26 27 28 29 30
$d
[1] 31 32 33 34 35 36 37 38 39 40
mapply(sum, list_1$a, list_1$b, list_2$c, list_2$d) [1] 64 68 72 76 80 84 88 92 96 100
applyfunctions are more computationally efficient than loops- you could also use
reshape2andplyr/dplyrpackages
aggregate
aggregate(age ~ lactation + da, data = health.wide, FUN = mean) lactation da age
1 1 No 2.235862
2 2 No 3.366667
3 3 No 4.417582
4 4 No 5.572917
5 5 No 6.668889
6 6 No 7.457143
7 7 No 8.816667
8 8 No 9.825000
9 9 No 10.100000
10 1 Yes 2.200000
11 2 Yes 3.750000
12 3 Yes 4.900000
13 4 Yes 5.600000
14 5 Yes 6.400000
15 7 Yes 7.900000
table
t1 <- table(health.wide$parity)
t1
1 2+
149 351
t2 <- table(health.wide$parity, health.wide$da)
t2
No Yes
1 145 4
2+ 339 12
prop.table(t1)
1 2+
0.298 0.702
prop.table(t2)
No Yes
1 0.290 0.008
2+ 0.678 0.024
prop.table(t2, margin = 2)
No Yes
1 0.2995868 0.2500000
2+ 0.7004132 0.7500000
See also xtabs(), ftable() or CrossTable() (in library gmodels).
Exercise 2
From the lesson on R objects, you created a matrix based on the following:
Three hundred and thirty-three cows are milked by a milker wearing gloves and
280 by a milker not wearing gloves. Two hundred cows in each group developed a
mastitis. Using the apply family of functions, find out the risk ratio and
odds ratio for mastitis according to wearing gloves.
| Mastitis | No mastitis | |
|---|---|---|
| Gloves | : 200 : | : 133 : |
| No gloves | : 200 : | : 80 : |
Solution
tab <- rbind(c(200, 133), c(200, 80))
rownames(tab) <- c("Gloves", "No gloves")
colnames(tab) <- c("Mastitis", "No mastitis")
row.tot <- apply(tab, 1, sum)
risk <- tab[, "Mastitis"] / row.tot
risk.ratio <- risk / risk[2]
odds <- risk / (1 - risk)
odds.ratio <- odds / odds[2]
rbind(risk, risk.ratio, odds, odds.ratio) Gloves No gloves
risk 0.6006006 0.7142857
risk.ratio 0.8408408 1.0000000
odds 1.5037594 2.5000000
odds.ratio 0.6015038 1.0000000
Split-Apply-Combine in Action
The following figure can give you a sense of what is split-apply-combine.
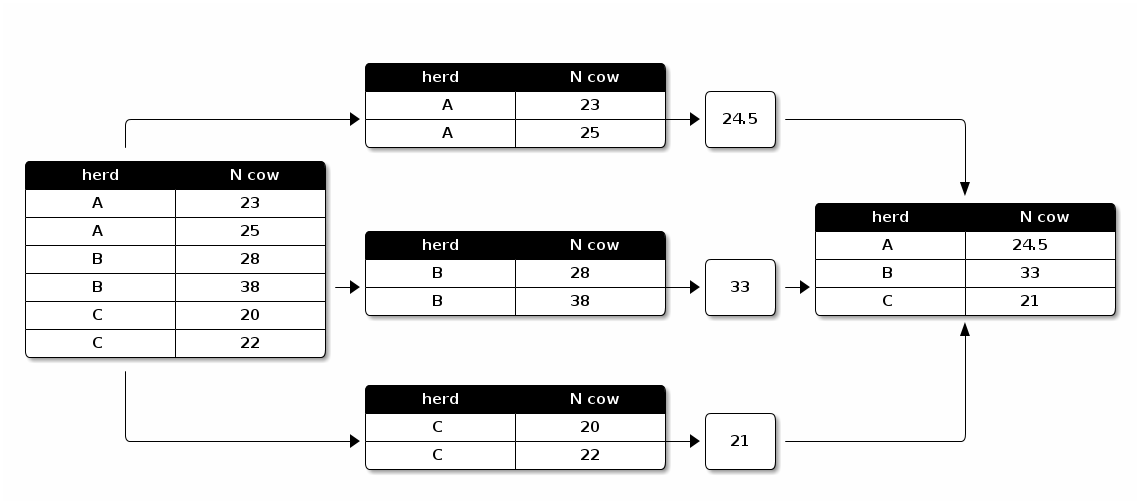
plyr package can be used to apply the split-apply-combine strategy. You need
to provide the following information:
- The data structure of the input
- The dataset being worked on
- The variable to split the dataset on.
- The function to apply to each split piece.
- The data structure of the output to combine pieces.
In short, plyr synthetizes the entire *apply family. For example, the mean
age by herd and parity could be computed and saved into a data frame:
lact <- ddply(health.wide,
.(herd, parity),
summarize,
mean.age = mean(age)
)Two other packages can also be helpful in applying this strategyL dplyr and
data.table.
